What is KAKA
KAKA is an integrative resource with detailed classification and annotation information for kinase activity related key alterations, which integrated 2553 data from literature curation and was divided into 4 categories (increase, decrease, kinase-dead, and no-effect), including 421 proteins in 8 species (Homo sapiens, Mus musculus, Saccharomyces cerevisiae, Rattus norvegicus, Arabidopsis thaliana, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe).
How to use KAKA
Quick Search
In the home page of KAKA, users can specify one field or all the fields to perform a quick search with the keyword(s) in the field(s). Users can simply click example and submit buttons to perform an example search.
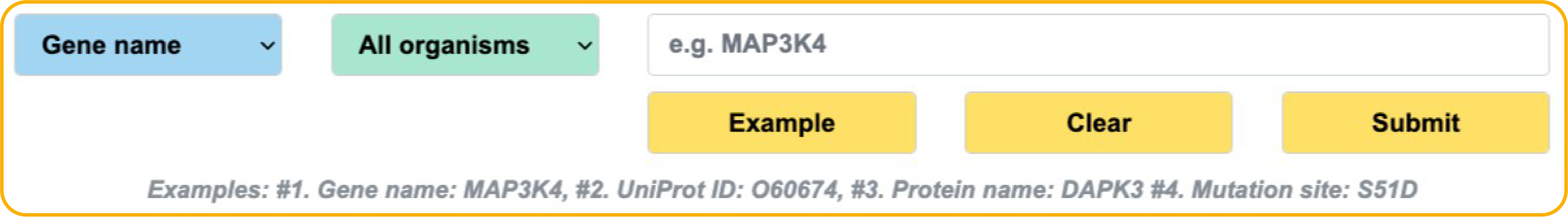
Advand search
In the Search page, users can add more keyword boxes and choose 'AND', 'OR' or 'NOT' to perform a more accurate multiple keywords search. Users can simply click example and submit buttons to perform an example search.
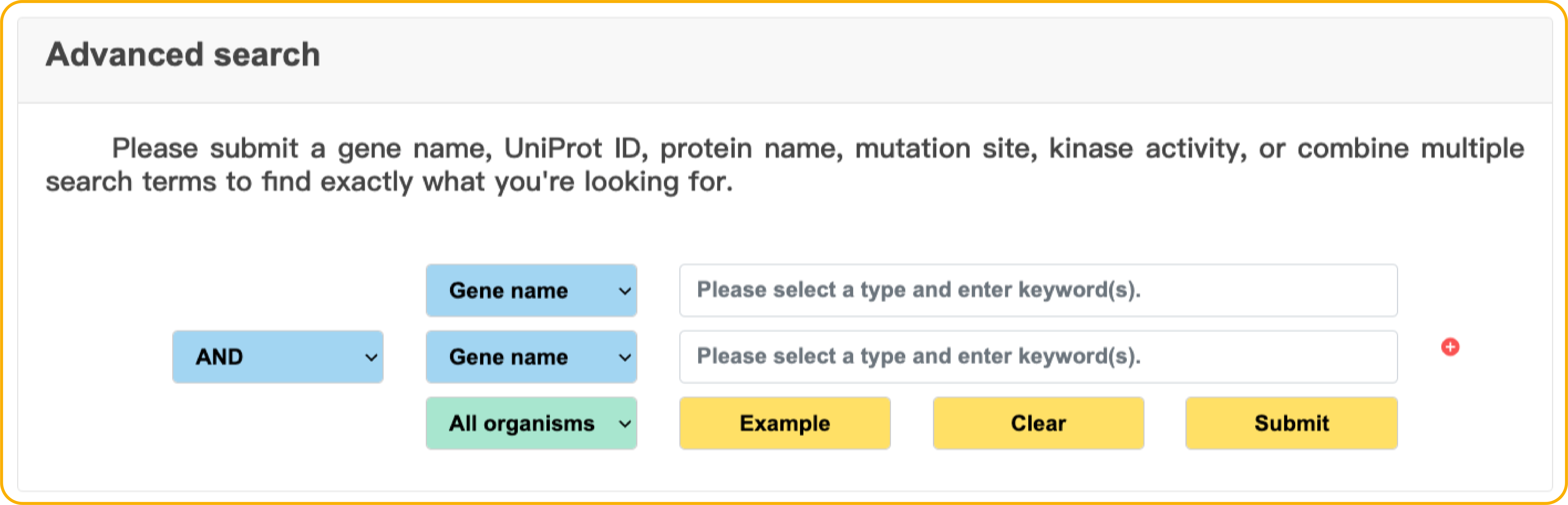
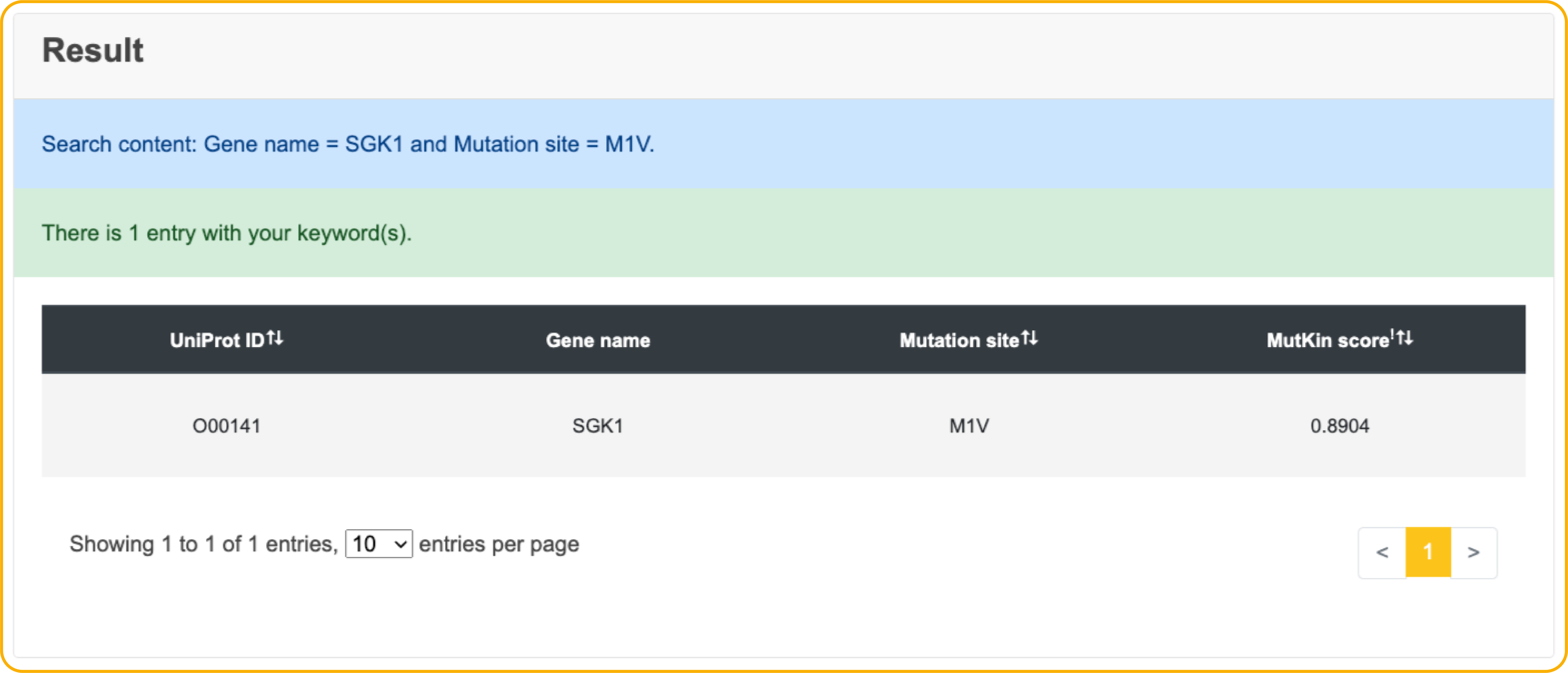
Search result
In the Result page, users can view the search or browse results, and further filter them by the options we provide. More information about experiment and protein is also displayed while users click the 'Detail' button.
Details
We manually collected the description of kinase activity from literature, and provided rich annotations for the kinase, such as protein sequence, kinase family, related functions of kinase genes and proteins and protein structure. The kinase substrate(s), kinase-specific phosphorylation sites, inhibitors(s) and mutation frequency are also available.

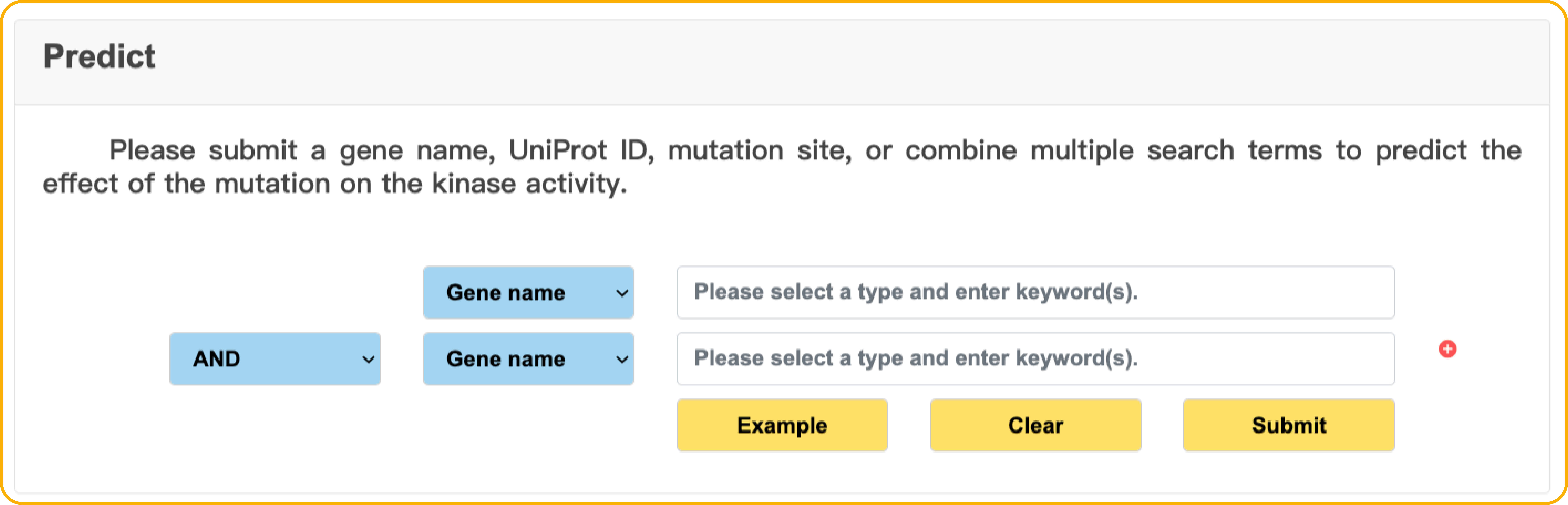
Predict
In the Predict page, we used KAKA to pre-computed predictions for all possible human kinase amino acid substitutions and missense variants.
Predicted result
Researchers can perform direct searches to obtain KAKA predictions.The prediction score was scaled between 0 and 1, representing the categorical probability of whether the mutation has an effect on kinase activity.
KAKA reference resources
UniProt
The mission of UniProt is to provide the scientific community with a comprehensive, high-quality and freely accessible resource of protein sequence and functional information.
https://www.uniprot.org/Gene Ontology (GO)
The Gene Ontology (GO) knowledgebase is the world’s largest source of information on the functions of genes. This knowledge is both human-readable and machine-readable, and is a foundation for computational analysis of large-scale molecular biology and genetics experiments in biomedical research.
http://geneontology.org/Kyoto Encyclopedia of Genes and Genomes (KEGG)
Kyoto Encyclopedia of Genes and Genomes (KEGG) is a knowledge base for systematic analysis of gene functions, linking genomic information with higher order functional information. The genomic information is stored in the GENES database, which is a collection of gene catalogs for all the completely sequenced genomes and some partial genomes with up-to-date annotation of gene functions. The higher order functional information is stored in the PATHWAY database, which contains graphical representations of cellular processes, such as metabolism, membrane transport, signal transduction and cell cycle.
http://www. genome.ad.jp/kegg/PhosphoSite
The experimentally identified downstream substrates and phosphorylation sites for the kinase were integrated from PhosphoSitePlus (http://www.phosphosite.org/), Phospho.ELM (http://phospho.elm.eu.org), RegPhos (http://csb.cse.yzu.edu.tw/RegPhos2/), HPRD (http://www.hprd.org), SWISS-PROT (http://www.ebi.ac.uk/swissprot/), and dbPTM (http://dbPTM.mbc.nctu.edu.tw/).
DrugBank
DrugBank is a web-enabled database containing comprehensive molecular information about drugs, their mechanisms, their interactions and their targets.
http://www.drugbank.caNetSurfP
NetSurfP-1,0, a tool for predicting solvent accessibility and secondary structure for each residue of an amino acid sequence using a feed-forward neural network architecture. The reliability of the surface accessibility prediction is stated in the form of a Z-score.
http://www.cbs.dtu.dk/services/NetSurfP/IUPred
Intrinsically disordered proteins and protein regions (IDPs/IDRs) exist without a single well-defined conformation. They carry out important biological functions with multifaceted roles which is also reflected in their evolutionary behavior. IUPred takes an amino acid sequence or a Uniprot ID/accession as an input and predicts the tendency for each amino acid to be in a disordered region with an option to also predict context-dependent disordered regions.
https://iupred3.elte.hu.The Cancer Genome Atlas (TCGA)
The Cancer Genome Atlas (TCGA) is a public funded project that aims to catalogue and discover major cancer-causing genomic alterations to create a comprehensive "atlas" of cancer genomic profiles. TCGA researchers have analysed large cohorts of over 30 human tumours through large-scale genome sequencing and integrated multi-dimensional analyses.
https://portal.gdc.cancer.gov/